
|
Secure genetic data moves into the fast lane of discovery
Take a ride down chromosome highways with a novel web-based platform called GWATCH that allows sharing of private genetic data while maintaining privacy through an ingenious -- and colourful -- dynamic visualization tool
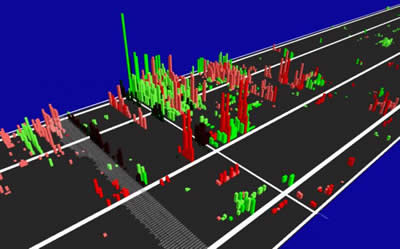
Image: This is an image capture from the dynamic 3-D Chromosome Highway Browser. Positive disease-associated regions are indicated by rising bars. Shown here is the CCR5 region on chromosome 3, which has been shown to be associated with HIV-AIDS
Photo Credit: Anton Svitin et al, GigaScience 2014, 3:18
November 5, 2014, Hong Kong, China - Today, the international open-access open-data journal GigaScience (a BGI and BioMed Central journal) announced publication of an article that presents GWATCH1, a
new web-based platform that provides visualization tools for identifying disease-associated genetic markers from privacy-protected human
data without risk to patient privacy. This dynamic online tool, developed by an international team of researchers from Russia,
Australia, Canada, and the US, allows and facilitates disease gene discovery via automation and presentation of intuitive
data visualization tools. GWATCH provides results in three dimensions via a scrolling (Guitar Hero-like) chromosome
highway. The reviewers get an extremely useful, visually appealing bird's-eye view of positive
disease-association results, while all sensitive information and raw data remain secure behind firewalls.
Identification of genes that underlie deadly complex diseases, such as heart disease, cancer and diabetes, and infections, including
HIV-AIDS, papilloma virus, and hepatitis B and C, is extremely difficult, as it requires the availability of a huge amount of genetic
information from large numbers of patients and healthy controls. The advent of cheaper and faster ways to sequence whole
genomes - with there likely to be over 200,000 human genomes sequenced this year2- has made producing this extensive
amount of data effectively a non-issue; however, issues over patient security and data access extremely limit
researchers' use of these amazing resources. Thus, identification of genes, replication of findings and
independent validation from 'potentially' available data is nearly impossible, due to the necessarily
complex and time consuming processes researchers need to go through to obtain access to protected
data. Thus, only a very small percentage of data in protected databases are ever used. To take
full advantage of these data to uncover ways to treat or prevent the ~20 million deaths per
year worldwide of people suffering from the most common complex diseases3, researchers
need new, secure methods to access and share these data.
Now, a large international collaboration of researchers from over 10 different institutions, led by Drs Anton Svitin and Stephen J. O'Brien,
developed a web-based tool called GWATCH (Genome-Wide Association Tracks Chromosome Highway), which does exactly this: allows access to
usable information from protected human data for discovery without revealing the underlying personal information or raw data.
One of the peer reviewers of the article, Lachlan Coin from the University of Queensland, made noted the importance of having such a tool,
saying "The discovery of novel genetic variants associated with complex disease has necessitated the formation of large global research
consortia to meta-analyse data from very large sample sizes. However, sharing of this data has always been problematic. GWATCH
provides an innovative web-platform to facilitate sharing of summary data from GWAS [Genome Wide Association Studies],
which will enable researchers to more quickly identify and validate disease-associated genetic variation."
GWATCH allows investigators who were not involved in the original study to access disease-associated genetic variation results from GWAS
(using whole genome sequence or SNP-arrays) rather than the raw data that can be used to identify individuals. GWATCH has a colourful and
dynamic, user-friendly visualization tool that enables researchers to effectively 'drive down chromosomes highways' and easily see
areas that associate with their disease of interest (See Figure). Further researchers can zoom in for greater detail on
variation patterns and see and compare different stages of disease (e.g., HIV infection, AIDS progression and
treatment outcome. A GWATCH tutorial video is available at http://www.youtube.com/watch?v=fIeOnZ-WLzo (and a just for fun music remix video at youtu.be/vNayRIk9fQA).
The authors developed and tested GWATCH using an often-requested huge dataset of association data from more than 6000 patients at risk
for HIV-AIDS, which had been previously collected by Dr O'Brien and colleagues with funding from the National Institutes of Health,
USA. GWATCH, however, can be used for any complex disease study by importing in that study's association results.
###
As part of GigaScience 's Open Science policy: the source code for GWATCH is freely available in Github4, an archived version of GWATCH
used in this paper is available in GigaDB5, and access to on-going updated versions of GWATCH is freely available
at http://gen-watch.org .
References:
1. Svitin A, Malov S, Cherkasov N, Geerts P, Rotkevich M, Dobrynin P, Shevchenko A, Guan L, Troyer J, Hendrickson-Lambert S, Hutcheson-Dilks
H, Oleksyk TK, Donfield S, Gomperts E, Jabs DA, Van Natta M, Harrigan PR, Brumme ZL, O'Brien SJ. GWATCH: a web platform for automated gene
association discovery analysis. GigaScience 2014, 3:18 http://www.gigasciencejournal.com/content/3/1/18
2. Regalado A. MIT Technology Review 2014. http://www.technologyreview.com/news/531091/emtech-illumina-says-228000-human-genomes-will-be-sequenced-this-year
3. World Health Organization. Top Ten Causes of Death 2012 http://www.who.int/mediacentre/factsheets/fs310/en/
4. https://github.com/DobzhanskyCenter/GWATCH
5. Svitin A, Malov S, Cherkasov N, Geerts P, Rotkevich M, Dobrynin P, Shevchenko A, Guan L, Troyer J, Hendrickson-Lambert S, Hutcheson Dilks H, Oleksyk TK, Donfield S, Gomperts E, Jabs DA, Van Natta M, Harrigan PR, Brumme ZL, O'Brien SJ. Software and supporting material for: GWATCH:
a web platform for automated gene association discovery analysis (2014) GigaScience Database http://dx.doi.org/10.5524/10.5524/100109.
GWATCH Tutorial Video: https://www.youtube.com/watch?v=fIeOnZ-WLzo GWATCH musical Remix: http://youtu.be/vNayRIk9fQA
This work was supported in part by Russian Ministry of Science Mega-grant 11.G34.31.0068 with Stephen J. O'Brien, Principal Investigator;
and by the National Institutes of Health, National Institute of Child Health and Human Development, R01-HD-41224.
Institutions Involved: Russia: Theodosius Dobzhansky Center for Genome Bioinformatics, St. Petersburg State University, St. Petersburg;
Department of Mathematics, St. Petersburg Electrotechnical University, St. Petersburg. Australia: Scientific Data Visualization
Consultant, Turner. USA: Genetics and Genomics Group, Advanced Technology Program, SAIC-Frederick, National Cancer
Institute, Frederick, MD; Department of Evolutionary Biology, Shepherd University, Shepherdstown, WV; Vanderbilt
Technologies for Advanced Genomics, Office of Research, Vanderbilt University Medical Center,
Nashville, TN; Biology Department, University of Puerto Rico, Mayaguez,
Puerto Rico; Department of Biostatistics, Rho, Inc., Chapel Hill,
NC; Division of Hematology-Oncology, Children's Hospital of Los Angeles, Los Angeles, CA; Departments of Ophthalmology and Medicine,
Icahn School of Medicine at Mount Sinai, New York, NY; Department of Epidemiology, The Johns Hopkins University Bloomberg School
of Public Health, Baltimore, MD; Oceanographic Center, Nova Southeastern University, Ft. Lauderdale, FL. Canada: British
Columbia Centre for Excellence in HIV/AIDS, Vancouver, BC; Division of AIDS, Faculty of Medicine, University of
British Columbia, Vancouver, BC; Faculty of Health Sciences, Simon Fraser University, Burnaby, BC
Media Contacts
Scott Edmunds
Executive Editor, GigaScience , BGI Hong Kong
Tel: +852 3610 3531
Mob: +852 92490853
Email: scott@gigasciencejournal.com
Notes to News Writers:
1. GigaScience is co-published by BGI, the world's largest genomics organization, and BioMed Central, the world's first open-access publisher. The journal covers research that uses or produces 'big data' from the full spectrum of the life sciences. It also serves as a forum for discussing the difficulties of and unique needs for handling large-scale data from all areas of the life sciences. The journal has a completely novel publication format - one that integrates manuscript publication with complete data hosting, and analyses tool incorporation. To encourage transparent reporting of scientific research as well as enable future access and analyses, it is a requirement of manuscript submission to GigaScience that all supporting data and source code be made available in the GigaScience database, GigaDB , as well as in their publicly available repositories. GigaScience can provide users access to associated online tools and workflows, and includes an integrated data analysis platform, GigaGalaxy, maximizing the potential utility and re-use of data. (Follow us on twitter @GigaScience; Facebook, and keep up-to-date on our blogs.
.
Source: http://www.eurekalert.org/pub_releases/2014-11/g-sgd110314.php
For more HIV and AIDS News visit...
Positively Positive - Living with HIV/AIDS:
HIV/AIDS News
|